2000-02-23: changing the initial orientation of the
In this series of calculations, the initial orientation of the ![]() group was along the crystallographic
group was along the crystallographic ![]() direction, i.e. along
the face diagonal of the cube of oxygen atoms inside the unit
cell. Moreover, relaxation of the defect was done without
simmetry (i.e. the keyword nodsymmetry was used).
direction, i.e. along
the face diagonal of the cube of oxygen atoms inside the unit
cell. Moreover, relaxation of the defect was done without
simmetry (i.e. the keyword nodsymmetry was used).
This calculations are to be compared with
2000-02-02 (3), in which the starting orientation of
the ![]() group was along the
group was along the ![]() direction and symmetry was
used for defect relaxation.
direction and symmetry was
used for defect relaxation.

Since here we change two variables (the nodsymmetry keyword
and the starting orientation of the ![]() group) a question
arises as to whether the nodsymmetry keyword makes any
difference when the same starting orientation of the
group) a question
arises as to whether the nodsymmetry keyword makes any
difference when the same starting orientation of the ![]() group is used.
group is used.
The following compares results for
![]() mixed
oxide using two input files which differ only for the presence
of the nodsymmetry keyword (the starting
mixed
oxide using two input files which differ only for the presence
of the nodsymmetry keyword (the starting ![]() orientation was
along the
orientation was
along the ![]() direction in both runs):
direction in both runs):
dschsun1:147> diff -c gd-OH gd-OH-sym *** gd-OH Wed Feb 23 12:39:55 2000 --- gd-OH-sym Thu Feb 24 10:53:50 2000 *************** *** 1,6 **** ! opti conp defect regi_before nodsymmetry #conp defect single ! dump every 1 gd-OH.dump maxcyc 200 cutb 3.0 # what does this mean? Ronny uses it. title --- 1,6 ---- ! opti conp defect regi_before #conp defect single ! dump every 1 gd-OH-sym.dump maxcyc 200 cutb 3.0 # what does this mean? Ronny uses it. title dschsun1:148>

Apparently, the only thing that changes is the total CPU time, which is more than doubled for the run which uses symmetry for defect relaxation. This is surprising, as I'd expect that use of symmetry would reduce CPU times.
Details for ![]() evaluation can be found in
2000-02-02 (3). Potential parameters are the same as that
calculation.
evaluation can be found in
2000-02-02 (3). Potential parameters are the same as that
calculation.
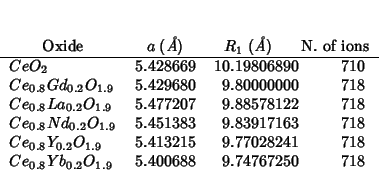

( Question: why are the CPU times so different from those in 2000-02-02?)
![]() values should be the same as 2000-02-02;
hence:
values should be the same as 2000-02-02;
hence:
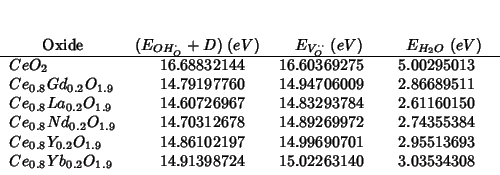
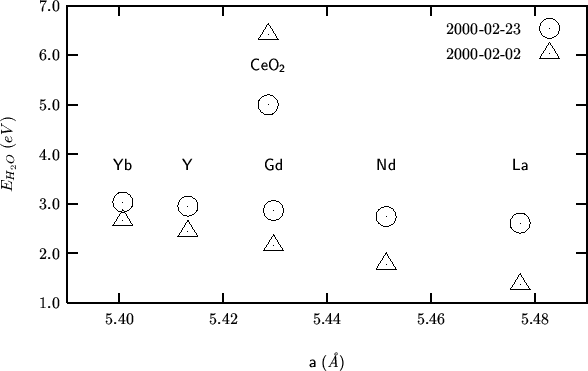
The trend is not changed with respect to 2000-02-02.
Differently from 2000-02-02, where the starting and optimized ![]() orientations are coincident, in this case the relaxed
orientations are coincident, in this case the relaxed ![]() orientation is along the
orientation is along the ![]() direction, i.e. along the body
diagonal of the cube defined by the eight oxygen ions inside the unit
cell.
direction, i.e. along the body
diagonal of the cube defined by the eight oxygen ions inside the unit
cell.
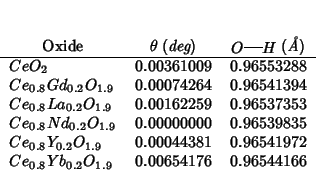
The following table shows the angle ![]() between the
between the
![]() vector and the
vector and the
![]() direction, and the
direction, and the
![]() bond length:
bond length:

opti conp defect regi_before nodsymmetry #conp defect single dump every 1 la-OH.dump maxcyc 200 cutb 3.0 # what does this mean? Ronny uses it. title Lanthanum solid solution N.Sakai et al. Solid State Ionics 125(1999)325-331 end cell 5.490000 5.490000 5.490000 90.000000 90.000000 90.000000 size 9.88578122 20.00 centre 0.25 0.25 0.25 impurity O2 core 0.25 0.25 0.25 interstitial H2 core 0.37354608 0.37354608 0.25000000 fractional ############ # Cores ############ Ce4 core 0.000000 0.000000 0.000000 -3.700000 0.8 La core 0.000000 0.000000 0.000000 3.00 0.2 O1 core 0.250000 0.250000 0.250000 0.07700 0.95 ############ # Shells ############ Ce4 shel 0.000000 0.000000 0.000000 7.700000 0.8 O1 shel 0.250000 0.250000 0.250000 -2.07700 0.95 space 225 ######################### # Shell-core charges ######################### species Ce3 shel 7.700000 Ce3 core -4.700000 O2 core -1.4263 H2 core 0.4263 ######################### # Short range potentials ######################### buck Ce4 shel O1 shel 1986.830000 0.351070 20.40000 0.0 15.000 buck Ce3 shel O1 shel 1731.61808 0.36372 14.43256 0.0 15.000 buck La core O1 shel 1439.7 0.36510 0.00000 0.0 15.000 buck O1 shel O1 shel 22764.300000 0.149000 27.89000 0.0 15.000 buck O2 core O1 shel 22764.300000 0.149000 27.89000 0.0 15.000 buck H2 core O1 shel 311.970 0.35 0.0 0.0 15.000 morse H2 core O2 core 7.05250 2.19860 0.9485 1.0 0.0 2.0 buck Ce4 shel O2 core 1986.830000 0.351070 20.40000 0.0 15.000 buck La core O2 core 1439.7 0.36510 0.00000 0.0 15.000 ######################### # Spring parameters ######################### spring Ce4 291.750000 spring Ce3 291.750000 spring O1 27.29